
Fusarium verticillioides, Fusarium graminearum and Ustilago maydis are diseases of the spigot of corn (Zea mays L.) that pose a risk to production. For this reason, a team of specialists from INTA Pergamino focuses on the study of the cereal genome to detect and identify common genes and metabolic pathways that are activated against these three pathogens. In the future, this finding will allow progress in the development of varieties with multiple resistance, capable of sustaining yields and improving the safety of the grain.
Since 2002, Juliana Iglesias, a specialist in plant genetics at INTA, has been advancing in the understanding of the molecular mechanisms that explain the resistance of corn to pathogens with a high productive and health impact. From a large-scale transcriptomic analysis, the research identified common genes associated with defense against spigot diseases that compromise both yield and grain quality.
The team coordinated by Iglesias adds strategic evidence to select candidate genes applicable to future genetic improvement programs. By identifying shared defensive routes against spigot diseases, the research opens the door to designing more resilient maize with greater productive stability.
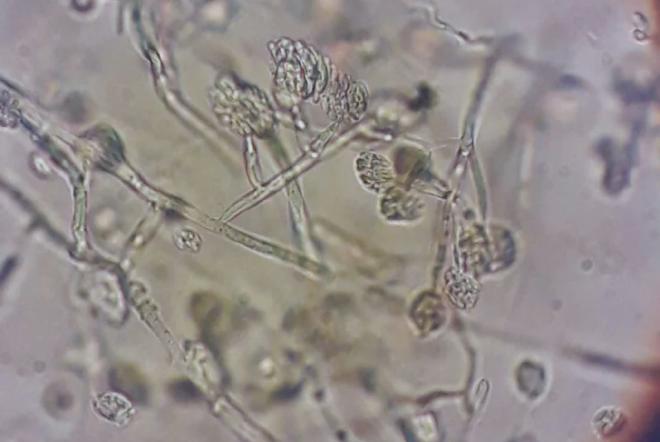
“With these results, we will be able to identify and study the genes that are activated in the response to multiple diseases to improve disease resistance in corn,” said the INTA biologist.
Andrea Peñas Ballesteros —within the framework of her master’s thesis in Bioinformatics and Systems Biology at the National University of the Northwest of the Province of Buenos Aires (UNNOBA)— focused on the interaction between corn and these three critical pathogens: Fusarium verticillioides and Fusarium graminearum cause rot that affects the filling of the spigot and, in addition, they generate mycotoxins – such as fumonisins and deoxynivalenol – that can enter the food chain. On the other hand, Ustilago maydis – which causes corn smut – severely alters the tissues of the spigot and reduces production, affecting the uniformity and commercial value of the crop.

The work takes on greater importance when understanding that maize DNA is made up of 32,000 genes inserted into 10 chromosomes. This confirms how complex the cereal genome is, because 85% of its genomic sequences are repeated multiple times.
Based on these data, candidate genes are being prioritised using a machine learning algorithm and, at the same time, they are being compared with previous Genome-Wide Association Study (GWAS) studies. Iglesias indicated that “an estimated 400 genes that could be associated with multiple resistance were classified and are being evaluated in functional field studies.”
Resistant genotypes presented a more effective and balanced defense response between defense and primary metabolism, maintaining cellular integrity and limiting infection. And susceptible genotypes showed a less effective response, with a metabolic conflict that prioritizes defense at the expense of growth and physiological stability.

The meta-analysis allowed to simultaneously compile, standardize and compare the responses of maize crops to these three pathogens with markedly differentiated pathogenesis styles.
“This finding opens up new opportunities for the genetic improvement of maize,” said Iglesias, while pointed out that “the precise knowledge of which genes are activated and how they interact against different pathogens allows us to accelerate breeding programs through the understanding of maize-pathogen interaction and selection assisted by molecular markers or even gene editing. A strategy that reduces time and costs.”
Citation #
- The article Maíz: identifican genes que ayudan a resistir enfermedades de la espiga was published on INTAinforma
Contact [Notaspampeanas](mailto: notaspampeanas@gmail.com)

