
“Most of the time, there are major limitations in the pathway to crop improvement,” said Iacopo Gentile, a postdoc in CSHL’s Zachary Lippman lab. “That’s because there’s so much redundancy and complexity in how gene families evolve and compensate for each other.”
“It’s about understanding what happens after gene duplication,” Gentile explained. “You have one gene that duplicates. Then you have two. What happens after that? Theory tells you they will diverge from each other. The big question mark in the field is how.”
To answer this question, the team homed in on CLE, a gene family involved in cell signaling and plant development. CLE peptides are prevalent across all plant species. However, much about their specific functions remains unknown. Studying them has been difficult due to their short length, rapid evolution, and redundancy.
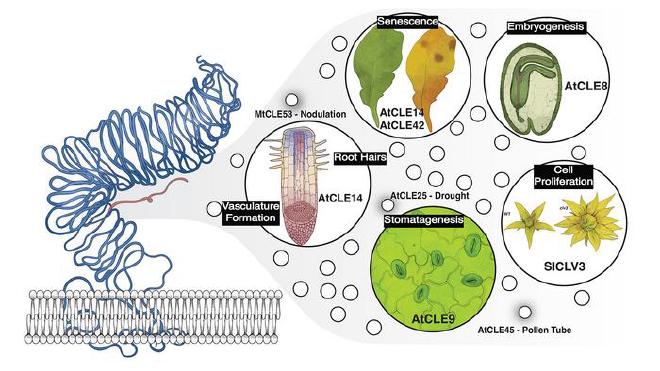
Using new advances in AI, the team identified thousands of previously unknown CLE genes across 1,000 species. They fed this data into computer models, which flagged genes that might be redundant. Redundant genes likely share similarities in one or two places—the peptides they produce, or gene promoters, the areas of DNA that control expression.

“It’s the first time in tomatoes where you have such big targeting of so many genes at the same time,” Gentile said. “We targeted 10.”
Gentile said the method they developed could “easily be scaled to every gene family,” not just CLE. As a result, plant breeders now have a “roadmap” to predict how hidden genes could be used to their advantage.
Citation #
- The study Pan-Angiosperm Analysis of the CLE Signaling Peptide Gene Family Unveils Paths, Patterns, and Predictions of Paralog Diversification was published on **Molecular Biology and Evolution**. Authors: Iacopo Gentile, Miguel Santo Domingo, Sophia G Zebell, Blaine Fitzgerald & Zachary B Lippman.
Acknowledgments #
The authors would like to thank members of the Lippman laboratory for their support and feedback and T. Mulligan, K. Schlecht, and S. Qiao for their assistance with plant care.
Funding #
Z.B.L. is supported by the National Science Foundation program grants IOS-2129189 and IOS-2216612 and the Howard Hughes Medical Institute.
Conflict of Interest #
Z.B.L. is a consultant for and a member of the Scientific Strategy Board of Inari Agriculture.
- The article AI offers ‘roadmap’ to plant genetics was published in CSHL’s website. Written by: Margaret Osborne, Science Writer | publicaffairs@cshl.edu
Contact [Notaspampeanas](mailto: notaspampeanas@gmail.com)

